onsite
onsite 🔬🎯
🚀 What is onsite?
onsite is a comprehensive Python package for mass spectrometry post-translational modification (PTM) localization. It provides algorithms for confident phosphorylation site localization and scoring, including implementations of AScore, PhosphoRS, and LucXor (LuciPHOr2).
✨ Key Features
- 🎯 Multiple Algorithms: AScore, PhosphoRS, and LucXor implementations
- 📊 Statistical Validation: Probability-based scoring with FLR estimation
- 💻 Unified CLI: Single command-line interface for all algorithms
- ⚡ Multi-threading: Parallel processing for improved performance
- 🔬 PyOpenMS Integration: Seamless integration with the OpenMS ecosystem
- 📈 High Accuracy: Confident site localization with statistical validation
- 🧩 Flexible API: Both command-line and Python API support
📊 Benchmark
We benchmarked onsite algorithms on the PXD000138 dataset using unified mzML/idXML inputs and consistent filtering (FDR < 0.01, tool-specific localization thresholds). The following results are obtained after applying algorithm-specific quality filters to ensure high-confidence site localization:
| Tool | Total PSMs | Total phospho sites | Well-resolved sites | Uncertain sites |
|---|---|---|---|---|
| LuciPHOr | 111,588 | 118,625 | 48,186 | 37,337 |
| AScore | 111,747 | 101,382 | 52,906 | 23,628 |
| pyLucXor | 111,588 | 117,341 | 51,468 | 38,970 |
| PhosphoRS | 111,747 | 107,552 | 50,000 | 26,354 |
Note: These counts represent phosphorylation sites from PSMs that passed both FDR filtering and tool-specific quality thresholds (local_flr < 0.01 for LuciPHOr/pyLucXor, AScore > 20 for AScore, site_prob > 99% for PhosphoRS).
FLR-Controlled Performance Comparison
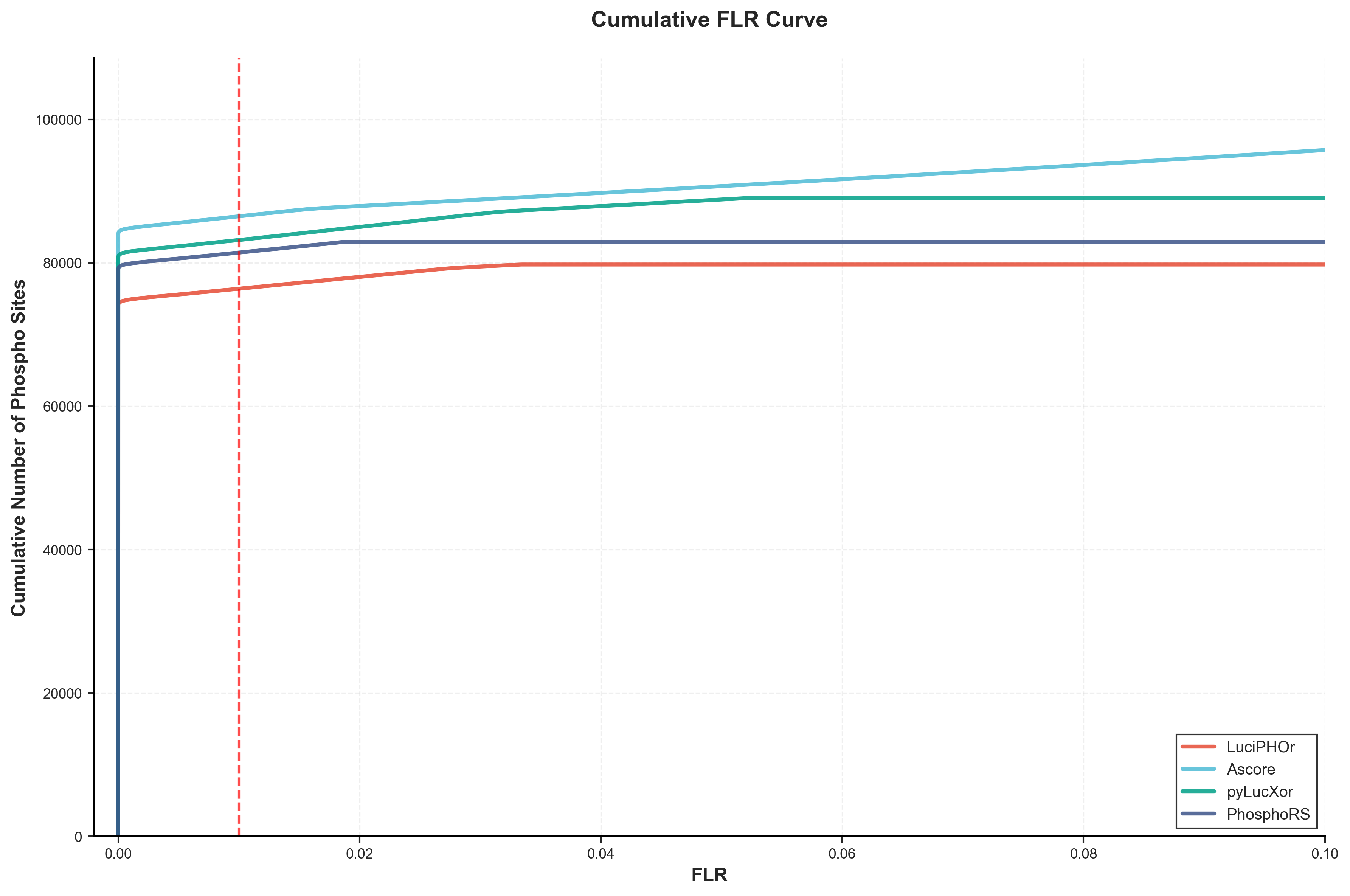
The cumulative FLR (False Localization Rate) curve demonstrates tool performance across different FLR thresholds without applying algorithm-specific filters, enabling unbiased cross-tool comparison. This analysis uses the pAla (phosphorylated Alanine) decoy strategy, where Alanine residues serve as decoy sites for FLR estimation, since Alanine cannot be biologically phosphorylated.
Phosphorylation Sites at Standard FLR Thresholds:
| Tool | Phospho_Count (1% FLR) | Phospho_Count (5% FLR) |
|---|---|---|
| LuciPHOr | 75,626 | 77,101 |
| AScore | 85,626 | 86,167 |
| pyLucXor | 82,349 | 84,397 |
| PhosphoRS | 80,618 | 81,473 |
At the recommended 1% FLR threshold, AScore identifies the most phosphorylation sites (85,626), followed by pyLucXor (82,349), PhosphoRS (80,618), and LuciPHOr (75,626).
See benchmark.md for methodology, full tables, and analysis details.
📋 Supported Algorithms
onsite provides three complementary algorithms for PTM localization:
1. AScore Algorithm
- Method: Probability-based approach using binomial statistics
- Features: Site-determining ion analysis, fast processing
- Output: AScore values indicating localization confidence
- Citation: Beausoleil et al. (2006) Nature Biotechnology
2. PhosphoRS Algorithm
- Method: Compomics-style scoring with isomer analysis
- Features: Site-specific probabilities, detailed isomer analysis
- Output: Site probability scores and isomer details
- Citation: Taus et al. (2011) Journal of Proteome Research
3. LucXor (LuciPHOr2) Algorithm
- Method: Two-stage processing with FLR estimation
- Features: False localization rate calculation, decoy-based validation
- Output: Delta scores, peptide scores, global and local FLR
- Citation: Fermin et al. (2013, 2015) MCP and Bioinformatics
💾 Installation
Prerequisites
- Python 3.11+
- PyOpenMS 3.5.0+
- NumPy 2.3.2+
- SciPy 1.16.1+
Using Poetry (Recommended)
# Clone the repository
git clone https://github.com/bigbio/onsite.git
cd onsite
# Install with Poetry
poetry install
# Activate the virtual environment
poetry shell
Using pip
# Install from PyPI (when available)
pip install onsite
# Or install from source
git clone https://github.com/bigbio/onsite.git
cd onsite
pip install -e .
Development Installation
# Clone the repository
git clone https://github.com/bigbio/onsite.git
cd onsite
# Install with development dependencies
poetry install --with dev
# Or with pip
pip install -e ".[dev]"
🛠️ Usage
Command Line Interface
onsite provides a unified command-line interface for all algorithms:
Unified onsite CLI
# AScore algorithm
onsite ascore -in spectra.mzML -id identifications.idXML -out results.idXML
# PhosphoRS algorithm
onsite phosphors -in spectra.mzML -id identifications.idXML -out results.idXML
# LucXor algorithm
onsite lucxor -in spectra.mzML -id identifications.idXML -out results.idXML
Individual Pipeline Tools
AScore Pipeline
# Basic usage
python -m onsite.ascore.cli -in spectra.mzML -id identifications.idXML -out results.idXML
# With custom parameters
python -m onsite.ascore.cli -in spectra.mzML -id identifications.idXML -out results.idXML \
--fragment-mass-tolerance 0.05 \
--fragment-mass-unit Da \
--threads 4 \
--add-decoys
PhosphoRS Pipeline
# Basic usage
python -m onsite.phosphors.cli -in spectra.mzML -id identifications.idXML -out results.idXML
# With custom parameters
python -m onsite.phosphors.cli -in spectra.mzML -id identifications.idXML -out results.idXML \
--fragment-mass-tolerance 0.05 \
--fragment-mass-unit Da \
--threads 1 \
--add-decoys
LucXor Pipeline
# Basic usage
python -m onsite.lucxor.cli -in spectra.mzML -id identifications.idXML -out results.idXML
# With custom parameters
python -m onsite.lucxor.cli -in spectra.mzML -id identifications.idXML -out results.idXML \
--fragment-method HCD \
--fragment-mass-tolerance 0.5 \
--fragment-error-units Da \
--threads 8 \
--debug
Command-line Options
AScore Options
| Option | Default | Description |
|---|---|---|
-in |
- | Input mzML file with spectra |
-id |
- | Input idXML file with identifications |
-out |
- | Output idXML file with scores |
--fragment-mass-tolerance |
0.05 | Fragment mass tolerance |
--fragment-mass-unit |
Da | Tolerance unit (Da or ppm) |
--threads |
1 | Number of threads for parallel processing |
--add-decoys |
False | Include decoy sites for validation |
--compute-all-scores |
False | Run all three algorithms and merge results |
--debug |
False | Enable debug logging |
PhosphoRS Options
| Option | Default | Description |
|---|---|---|
-in |
- | Input mzML file with spectra |
-id |
- | Input idXML file with identifications |
-out |
- | Output idXML file with scores |
--fragment-mass-tolerance |
0.05 | Fragment mass tolerance |
--fragment-mass-unit |
Da | Tolerance unit (Da or ppm) |
--threads |
1 | Number of threads for parallel processing |
--add-decoys |
False | Include decoy sites for validation |
--compute-all-scores |
False | Run all three algorithms and merge results |
--debug |
False | Enable debug logging |
LucXor Options
| Option | Default | Description |
|---|---|---|
-in |
- | Input mzML file with spectra |
-id |
- | Input idXML file with identifications |
-out |
- | Output idXML file with scores |
--fragment-method |
CID | Fragmentation method (CID or HCD) |
--fragment-mass-tolerance |
0.5 | Fragment mass tolerance |
--fragment-error-units |
Da | Tolerance units (Da or ppm) |
--min-mz |
150.0 | Minimum m/z value to consider |
--target-modifications |
Phospho (S/T/Y) | List of target PTM definitions |
--neutral-losses |
sty -H3PO4 -97.97690 | Neutral loss definitions applied during scoring |
--decoy-mass |
79.966331 | Mass offset used when generating decoy permutations |
--decoy-neutral-losses |
X -H3PO4 -97.97690 | Neutral loss patterns for decoy permutations |
--max-charge-state |
5 | Maximum charge state |
--max-peptide-length |
40 | Maximum peptide length |
--max-num-perm |
16384 | Maximum permutations |
--modeling-score-threshold |
0.95 | Minimum score for selecting PSMs during model building |
--scoring-threshold |
0.0 | Minimum LucXor score to report |
--min-num-psms-model |
50 | Minimum number of high-scoring PSMs required for modeling |
--threads |
1 | Number of threads for parallel processing |
--rt-tolerance |
0.01 | RT tolerance used when matching spectra by retention time |
--disable-split-by-charge |
False | Disable splitting PSMs by charge state for model training |
--compute-all-scores |
False | Run all three algorithms and merge results |
--debug |
False | Enable debug logging |
📊 Algorithm Details
AScore Algorithm
The AScore algorithm provides phosphorylation site localization by analyzing MS/MS fragment ions to identify site-determining ions and computing localization probabilities based on fragment evidence.
Output Metrics:
AScore_pep_score: Overall peptide scoreAScore_1, AScore_2, ...: Individual site scoresProForma: Standardized sequence notation with confidence scores
PhosphoRS Algorithm
The PhosphoRS algorithm implements a comprehensive approach using isomer generation, theoretical spectrum matching, and probability scoring for confident phosphorylation site assignment.
Output Metrics:
- Site-specific probability scores (0-100%)
- Isomer details with sequence and score
- Detailed confidence metrics
LucXor (LuciPHOr2) Algorithm
LucXor implements the complete LuciPHOr2 algorithm with two-stage processing for accurate PTM localization with false localization rate (FLR) estimation.
Output Metrics:
Luciphor_delta_score: Main localization scoreLuciphor_pep_score: Peptide identification scoreLuciphor_global_flr: Global false localization rateLuciphor_local_flr: Local false localization rate
🔍 Example Results
You can find example result files in the data directory. Here are the direct links to different algorithm result files:
| Algorithm | Description | Result File |
|---|---|---|
| AScore | AScore phosphorylation site localization results | AScore Example |
| PhosphoRS | PhosphoRS phosphorylation site localization results | PhosphoRS Example |
| LucXor | LucXor (LuciPHOr2) PTM localization results with FLR | LucXor Example |
📖 Documentation
For more detailed information:
- AScore Algorithm Documentation
- PhosphoRS Algorithm Documentation
- LucXor Algorithm Documentation
- Citations and References
👥 Contributing
To contribute to onsite:
- 🍴 Fork the repository
- 📥 Clone your fork:
git clone https://github.com/YOUR-USERNAME/onsite - 🌿 Create a feature branch:
git checkout -b new-feature - ✏️ Make your changes
- 🔧 Install in development mode:
pip install -e . - 🧪 Test your changes:
poetry run pytest - 💾 Commit your changes:
git commit -am 'Add new feature' - 📤 Push to the branch:
git push origin new-feature - 📩 Submit a pull request
📜 License
This project is licensed under the MIT License - see the LICENSE file for details.
📝 Citation
If you use onsite in your research, please cite:
onsite: Mass spectrometry post-translational modification localization tool.
https://github.com/bigbio/onsite
🔗 Related Tools
- PyOpenMS - Python bindings for OpenMS
- OpenMS - Open-source tools for mass spectrometry
- nf-core/quantms - Quantitative mass spectrometry workflow
❓ Need Help?
If you have questions or need assistance:
- Open an issue on GitHub
- Check existing issues for solutions
🙏 Acknowledgments
onsite builds upon the excellent work of the original algorithm developers and the OpenMS community. We thank all contributors and users for their feedback and support.



